Check the dispersion of a model
Thierry Onkelinx
2019-01-21
Source:vignettes/dispersion.Rmd
dispersion.RmdGenerate data
library(inlatools)
dataset <- generate_data()Fit the models
library(INLA)
#> Loading required package: Matrix
#> Loading required package: sp
#> This is INLA_18.07.12 built 2018-07-20 09:17:00 UTC.
#> See www.r-inla.org/contact-us for how to get help.
#> To enable PARDISO sparse library; see inla.pardiso()
poisson_model <- inla(
poisson ~ 1 + f(group_id, model = "iid"), family = "poisson",
data = dataset,
control.compute = list(config = TRUE, waic = TRUE)
)
negbin_model <- inla(
negbin ~ 1 + f(group_id, model = "iid"), family = "poisson",
data = dataset,
control.compute = list(config = TRUE, waic = TRUE)
)
binom_model <- inla(
binom ~ 1 + f(group_id, model = "iid"), family = "poisson",
data = dataset,
control.compute = list(config = TRUE, waic = TRUE)
)Test the dispersion
plot(dispersion_check(poisson_model))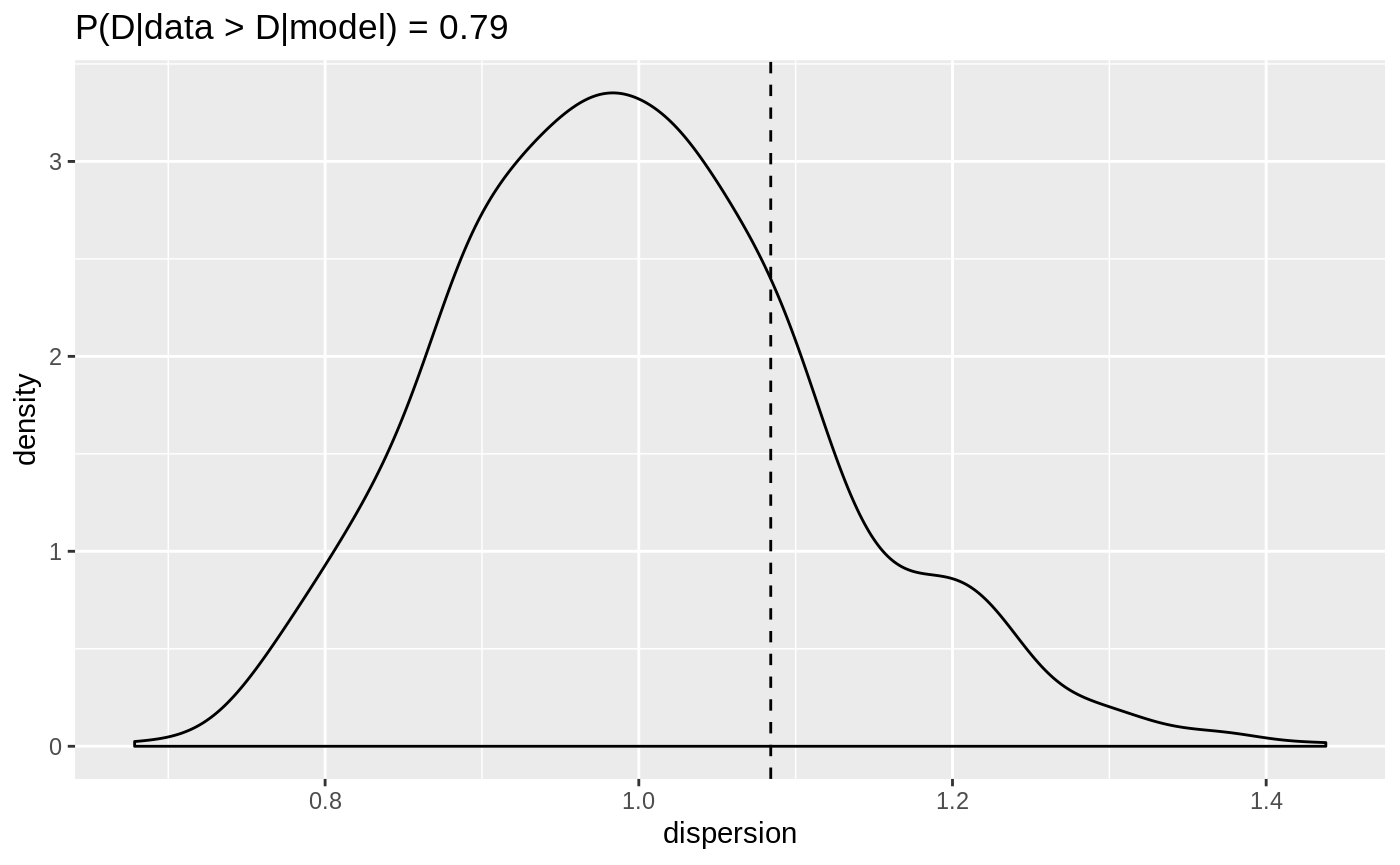
Dispersion of the Poisson response
plot(dispersion_check(negbin_model))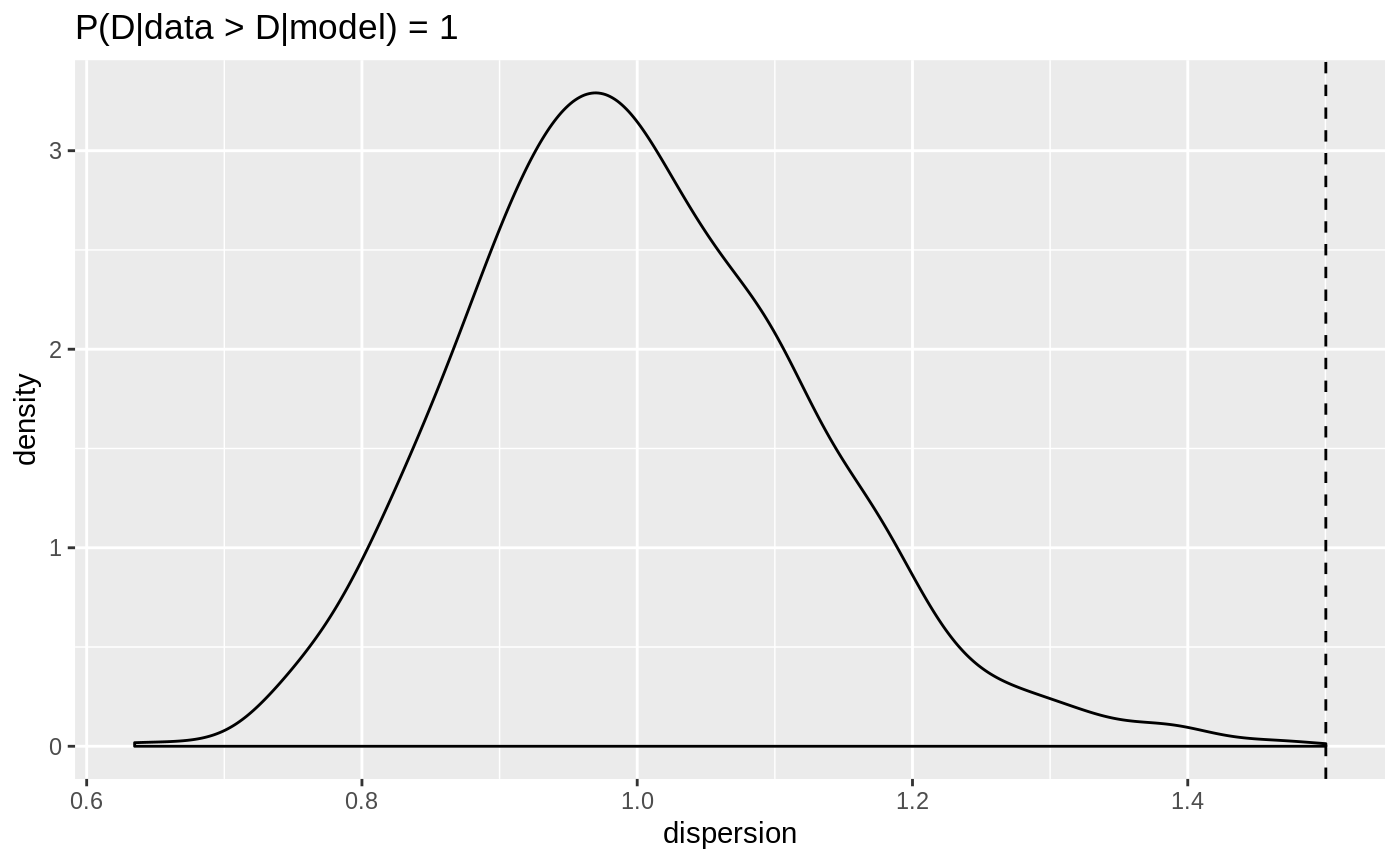
Dispersion for the negative binomial response
plot(dispersion_check(binom_model))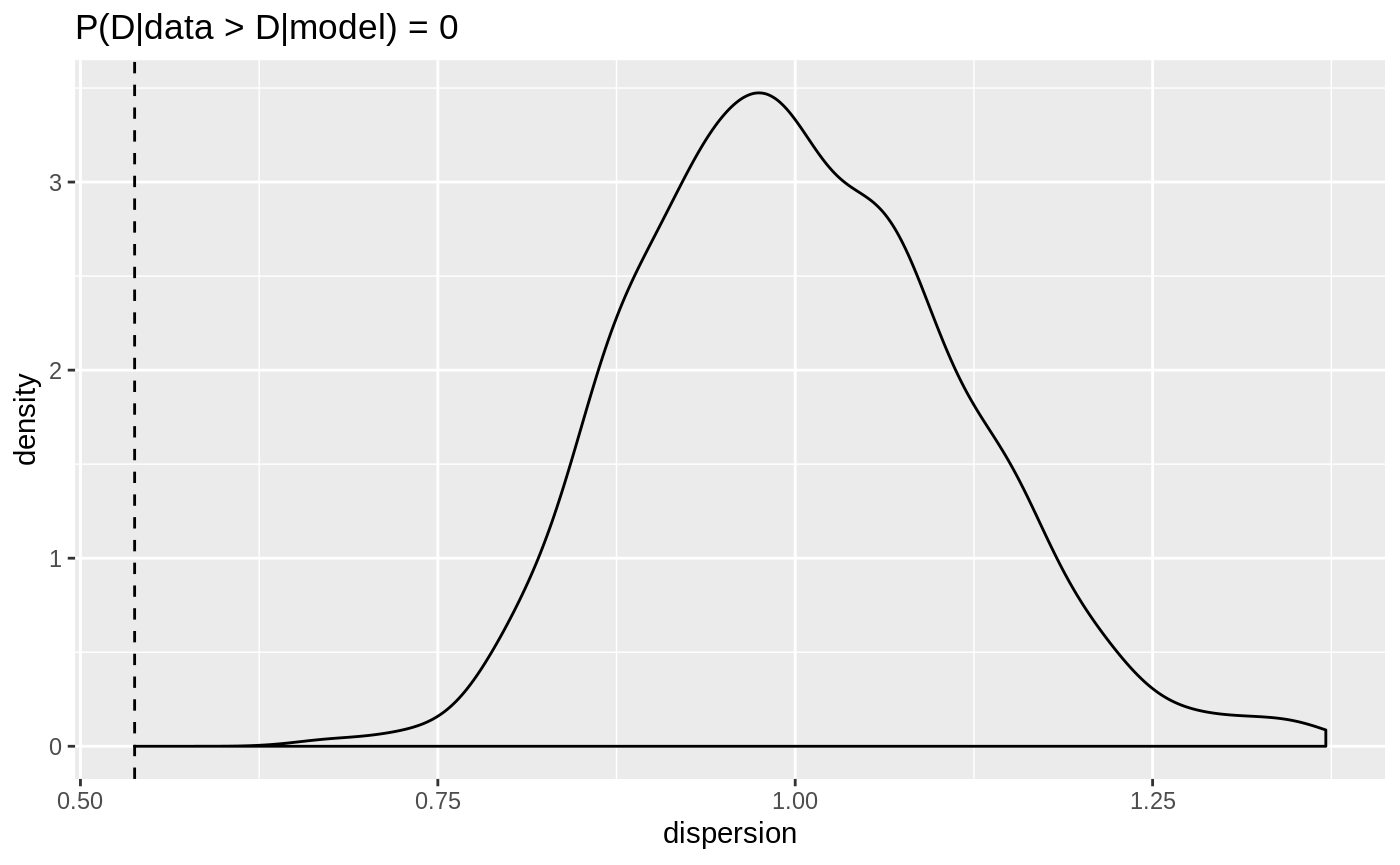
Dispersion for the negative binomial response
Simulation study
The simulation study below is not run because it takes some time to run.
n_replicate <- 100
nsim <- 100
dispersion_poisson <- function(i, nsim = 100) {
dataset <- generate_data()
model <- inla(
poisson ~ 1 + f(group_id, model = "iid"), family = "poisson",
data = dataset,
control.compute = list(config = TRUE, waic = TRUE)
)
D <- dispersion_check(model, nsim = nsim)
mean(D$data > D$model)
}
dispersion_negbin <- function(i, nsim = 100) {
dataset <- generate_data()
model <- inla(
negbin ~ 1 + f(group_id, model = "iid"), family = "poisson",
data = dataset,
control.compute = list(config = TRUE, waic = TRUE)
)
D <- dispersion_check(model, nsim = nsim)
mean(D$data > D$model)
}
dispersion_binom <- function(i, nsim = 100) {
dataset <- generate_data()
model <- inla(
binom ~ 1 + f(group_id, model = "iid"), family = "poisson",
data = dataset,
control.compute = list(config = TRUE, waic = TRUE)
)
D <- dispersion_check(model, nsim = nsim)
mean(D$data > D$model)
}
sim_dispersion_poisson <- map_dbl(
seq_len(n_replicate),
dispersion_poisson,
nsim = nsim
)
sim_dispersion_negbin <- map_dbl(
seq_len(n_replicate),
dispersion_negbin,
nsim = nsim
)
sim_dispersion_binom <- map_dbl(
seq_len(n_replicate),
dispersion_binom,
nsim = nsim
)
library(dplyr)
library(ggplot2)
data.frame(
response = rep(
c("Poisson", "negative\nbinomial", "binomial"),
each = n_replicate
),
proportion = c(
sim_dispersion_poisson, sim_dispersion_negbin, sim_dispersion_binom
)
) %>%
ggplot(aes(x = proportion)) +
geom_density() +
facet_wrap(~response, scales = "free_y")